Frequently Asked Questions
Published:
… although some are not frequently asked, but might nevertheless be useful. Below questions and answers in random order. Please be sure to check out the official list of FAQ first. Should you have other questions not appearing on either page, please create a new issue on Github, ask the question on BlueSky, or send an email to the AEA Data Editor.
Citing, DOI
What is the DOI of my openICPSR deposit? I have not yet published it, but am asked to add a citation to it in my manuscript?
Generically, each openICPSR project has a number (e.g., “109622”), that might show up on the right panel:
 Then
Then
- if the openICPSR project has not been published, then the DOI will be “http://doi.org/10.3886/E” + number + “V1” (e.g. http://doi.org/10.3886/E109622V1)
- if the openICPSR project has already been published, then the CURRENT DOI is shown on the relevant page, but if there are any revisions the Data Editor has asked for, then the to-be-cited DOI would be the next version, e.g., “http://doi.org/10.3886/E” + number + “V3” if the current version is V2 and the next version would be V3.
Give it a try:
How do I cite my own data and code supplement?
If you created your own data (experiments, surveys, etc.), you should do one of two things:
- If you believe that you will re-use the data as-is, and in particular if you would like others to also use the data, we strongly suggest creating a separate data deposit at a data repository (see this guidance at the Social Science Data Editor website. This deposit does not need to be at the AEA Data and Code Repository - it can be at any trusted repository. Have a look at the Social Science Data Editor’s guide to Data and Code Hosting. Once deposited, and published, the data should be cited in your manuscript, in accordance with the AEA Sample References.
- If you only intend to write this one manuscript with the data, you should cite the manuscript’s companion data and code deposit at the AEA Data and Code repository. Since that deposit is (typically) not yet published, you need to manually construct the reference, as per the AEA Sample References (an example is given there). The DOI for your forthcoming data and code deposit can be constructed as outlined in the previous FAQ.
I have a paper that uses data from 14 different sources. How do I comply with requirement for data citations and fit within page limits? (for instance in Papers and Proceedings).
We understand page limits, here are possible workarounds, in decreasing order of preference:
- collect “related” citations into a consolidated citation. This should NOT simply be the citation for the data provider, but a meaningful collection. See this example.
- refer to an (online) appendix for the details, and have an appendix-only bibliography exhaustively listing all sources (may not be an option for Papers and Proceedings)
- refer to the README in the deposit, which has a bibliography like a real paper (see the README template).
Note that citing the sources is a formal requirement. You may still want to provide a table with a more compact list of links or series identifiers, for quick reference or machine-readability.
Licensing
Are authors allowed to reuse the data once the data is published on the AER webpage and what are the specific conditions?
As of Jan 2021, this question has two answers, pending final migration of archival replication packages:
If the deposit is still downloaded from the AEA website (download URL starts with “https://www.aeaweb.org”)
The copyright of older deposits was transferred (© AEA), together with the manuscript, to the AEA. However, we encourage re-use, and permission is automatically granted to any user of these deposits to use and re-publish them, under acknowledgement (citation) of the authors original paper and replication package. See sample references, example no. 4.
If the deposit is downloaded from the AEA Data and Code Repository at openICPSR (download link redirects to a page at the Repository)
Migrated deposits are (© AEA), newer deposits remain copyrighted by the original authors (unless otherwise stated in the deposit’s LICENSE file). Most deposits are under a mixed CC-BY/BSD license, or under a CC-BY license.

Both licenses allow for re-use and re-distribution, under acknowledgement (citation) of the DOI of the replication package, see sample references, example no. 3.
Deposit related
Should we keep the data and directory structure as we used it ourselves or should we set up the files in a way that would make replication as straightforward as possible?
… the directory structure has gotten a little clunky over the years working on this project…
The Data and Code Availability Policy says:
“Files uploaded to the AEA Data and Code Repository should retain the file names as originally executed or used, their original file format, and their original “grouping” in terms of directories.”
You should feel free to reorganize, but you should ensure when we run the reorganized files, they produce the same results that are reported in the paper. Or put differently, the numbers in the paper should be produced by the reorganized files. We are not trying to reproduce your historical path to the paper, only the current state of the paper.
Such restructuring may also be appropriate if you have a very sophisticated reproducible setup in your lab or group. A replicator does not need all sorts of fancy dynamic setup scripts that are very relevant in a lab, but unnecessarily complicate the process for a replicator. You should attempt to simplify the final setup to make it easy for anybody to run this particular project, once.
I was asked to modify files in my repository (not yet published) but I cannot upload or edit anything
When you first submitted to the AEA, your deposit became locked. There are two ways it can be edited:
You can “recall” the submission
On the right, under “ Change status”, choose “Recall submission”
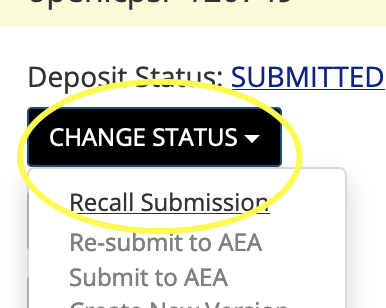
You should then be able to upload and make changes.
Once you are done, choose “Re-submit” from the same menu.
The Data Editor staff can request revisions
If you received a notice via the openICPSR communication log requesting revisions, you should be able to make modifications as outlined in the request. You should be all set.
Again, once you are done, choose “Re-submit” from the same menu as above.
I was wondering whether (and how) I can update the published repository for our paper. I was contacted by a researcher who is doing a replication … couple of minor issues … forgotten to include two auxiliary datasets in the repository without which one of the programs does not run successfully.
First off, excellent initiative. Our team cannot always conduct a full replication (not all data may be accessible, not enough time, no access to the software). We appreciate it when others are able to do that work, and when authors then correct the replication package.
Updating the repository is actually very easy, and updates likes these are exactly why we moved to the openICPSR repository for this. We have a policy how changes are then recorded, see https://www.aeaweb.org/journals/data/policy-revisions.
1) Log back onto your openICPSR deposit. If you don’t remember, simply click on the “Share Data” link on openICPSR, and it will show you your deposits.
2) You will need to click on “Create new version” if you created the deposit after after July 2020).

3) Update the README as per the policy. The updated README should have a section called “Changes” (or a separate file called “CHANGES.txt”). Authors should list the files added, any changes made to the programs, and ideally the reason why. No more than a paragraph per version, e.g.,
The data and code in this deposit have been updated after publication of the article.
- V1: Original version
- V2: The code has been simplified, and better instructions provided. All results are the same.
- V3: Permission was obtained by the data provider to post additional data. “Master.do” and the instructions in the README have been updated. Figures 5, 8, and 10 are now reproducible with this archive.
4) Once you updated all files (remember to update the README), choose “Submit to AEA” in “Change Status”. Please also notify the Data Editor by email, separately.
5) The AEA Data Editor will review that the criteria of the Revision Policy are satisfied, but conduct no other checks.
6) In most cases, the article will remain linked to the V1 deposit (“version of record”), but anybody navigating there will see a banner indicating that a more recent version exists (the V2 deposit).

I am trying to upload to the repository, but my replication package has more than 1,000 files
As of 2021, the 1,000 file limit is unfortunately a hard limit. Therefore, we relax the rule that all data and code should be unzipped, though we still insist on the “smallest possible configuration”.
In most cases, it is a particular directory that is the primary culprit. Say you have
Structure-pre:
/code: 20 files
/data/
src1/: 25 files
src2/: 101 files
src3/: 3,000 files
then the ideal structure, taking into account the 1,000 file limit, would be:
Structure-post:
/code: 20 files
/data/
src1/: 25 files
src2/: 101 files
src3.zip: = 1 file, containing 3,000 files
(src3/ and its 3,000 files have been removed!)
Your README should provide instructions to the replicators how to recover the fully unzipped structure (there are cross-platform differences in unzipping, so be precise about the final structure, rather than the method of getting there).
Alternatively, the code can handle the unzipping - optional, but more robust.
Once you’ve adjusted that, zip up the whole structure (so a ZIP file that has inside it another zip file, plus the /code, /data/src1/, and /data/src2/ directories), and “Import from ZIP” when uploading to ICPSR.
For an example on how to document this in your repository, see this file. You can incorporate such text into a separate file, into the README (preferred), or adjust your code to take this structure into account (much preferred).
What is that __MACOSX folder, which seems to contain a second copy of all the replication files (I am not sure why this folder exists)
[MAC USERs ONLY] We are also not sure, but it is a standard feature of ZIP files created on Mac OSX systems using the graphical user interface. Here’s a quick fix that helps all parties involved (adapted from this source):
- Create your ZIP file as usual
- Open the Terminal App
- Start typing
zip -d(note space) - Drag the ZIP file onto the Terminal
- Complete the command line with ` “__MACOSX*”` and hit enter.
The whole thing should look like this:
$ zip -d /Users/myname/Workspace/Folder/myzip.zip "__MACOSX*"
deleting: __MACOSX/
deleting: __MACOSX/myzip/
deleting: __MACOSX/myzip/._Proof_hi.pdf
deleting: __MACOSX/myzip/._README.pdf
deleting: __MACOSX/._myzip
You can now upload the file to openICPSR using the “Import from ZIP” functionality.
We should note that these folders do not show up in the public view of the repository once it is published. So while it is probably OK to leave them, it is better to remove them.
Confidential data
The paper uses confidential data, covering [geography] for period [2001-2015]. The repository only contains code. Should the repository metadata be filled out for the data characteristics, even if the repository only has code?
[Answer from ICPSR] I think it still makes sense to complete as much metadata as possible. There are syntax files specific to the data available through a restricted-use agreement. The metadata are for increasing findability of the data collection – even if only the syntax are in the repository. It’s useful to know the data analyzed with the syntax are about a specific geographic coverage for a specific time period.
I use confidential data. I am allowed to provide the data to the Data Editor for the purpose of replication, but you are not allowed to publish the data. How do I proceed?
I have been told by the Data Editor to remove PSID data from my submitted materials. What do I do?
Version control, Software, etc.
We used custom software. How do we describe/provide access to that?
This case might arise if authors ran an experiment or a novel type of survey, using software specifically created for the task. Our policy does require
“information about […] details of the computations sufficient to permit replication,”
and requires that “programs” be archived at the AEA Data and Code Repository. Furthermore,
“replication materials shall also include […] (g) computer code for experiment or survey collection mechanisms.”
However, we do not require that software, as opposed to code and programs that configure and instruct the software, be uploaded.
- Software: a “collection of instructions and data that tell a computer how to work” [1], here best approximated by “application software”, “a computer program designed to carry out a specific task other than one relating to the operation of the computer itself.” Stata, Matlab, Qualtrics, zTree are software for the purpose of the policy, as are compilers such as Intel or GNU Fortran.
- Code or programs here refers to instructions to the application software to produce a specific output - instructions that “allow end-users – people who are not professional software developers – to program computers.” [2]. Stata code, Qualtrics configuration files, and to some extent Fortran source code are typically considered to be “code.”
Much of economics research is conducted using proprietary software - Stata, Matlab, etc. Our policy requires that the software be available for use by others - possibly paying a fee - and that any configuration parameters (“code”) be available. For experiments, this might mean providing zTree code, but not the software itself (which is subject to a license agreement); for surveys, this might mean providing Qualtrics configurations, but not access to a Qualtrics subscription. We do not typically require information about how to purchase or download such frequently used, named software. In most cases, the software licenses prohibit redistribution, but even in the case of open-source software (R, Julia, Python, etc.), we do not require that authors upload the executable or source code.
In other cases, the software might have been custom-designed for the purposes of the authors’ research. In that case, the README should contain information on how to obtain the software. If the software is under an open source license, this might entail providing a link to a website and the software’s installation and compilation instructions. If the software is not under an open source license, the README might contain language such as
“We contracted with (DEVELOPER) to develop the software used to conduct the experiment, at a cost of approximately (rounded to next $100 or $1000). To obtain the software, (DEVELOPER) can be reached at (EMAIL) or (WEBSITE).”
If software is not open source, then two cases may occur:
- If the developer retains ownership of the software, then we are talking about a licensing agreement, much like when a researcher purchases a yearly or permanent license to Stata or Matlab. No further information is required.
- However, if the authors contracted with the developer of the software, the authors may also have rights to the software.
- If developer and authors co-own the software, the README should state what rights authors confer automatically to any replicator. For instance, if the developer needs the authors’ approval to license (at some price) to any other researcher, then the README should explicitly state that the authors provide such approval.
- If the authors obtained full rights to the end product (a true purchase), then they should provide a mechanism to obtain the software, and state explicitly again what rights replicators have to use the software. This might include demonstration modes sufficient to conduct the replication.
We already use git/svn/GitHub/GitLab/BitBucket/etc. Do you facilitate integration of existing version-controlled code to the AEA repo? Or even planned functionality for linking out directly to such projects where they can be found online?
Some econometrics papers might be accompanied by (for example) an R or Stata package (perhaps published on CRAN or SSC). What about surfacing references to associated packages more prominently?
Do you support Docker/ Jupyter/ etc.?
RCT
Aligning AEA RCT Registry and AEA Data and Code Repository
The AEA RCT registry has a field that codes whether data associated with a registration is publicly available. Many authors will have this coded as “non public” prior to the publication of the replication package. When the replication package is about to be published on the AEA Data and Code Repository, this field needs to be updated. Only the authors of the registry can update this field. Steps to follow:
- Log in to the AEA RCT registry and select your registration
- Change the field to “public” / “published”
- Compute the DOI of your forthcoming replication package publication and enter the resulting DOI in the URL field.
- Do not use the URL of the openICPSR project in the browser address bar!
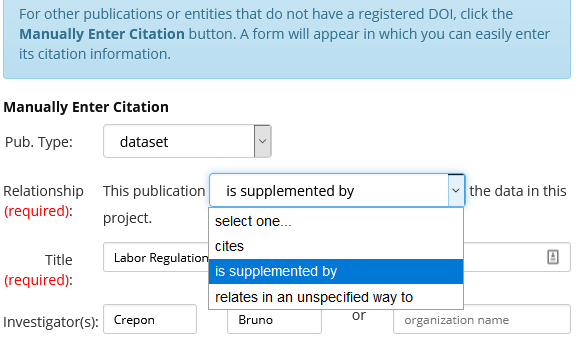
[EXTRA] You should also record the RCT DOI as a related publication of your deposit on the AEA Data and Code Repository:
- The RCT registry will show the DOI of your registration at the bottom of its public page. Example:

- You can then enter that DOI (e.g.,
10.1257/rct.156-1.1) into the “Related Publication” field of the deposit on the AEA Data and Code Repository:
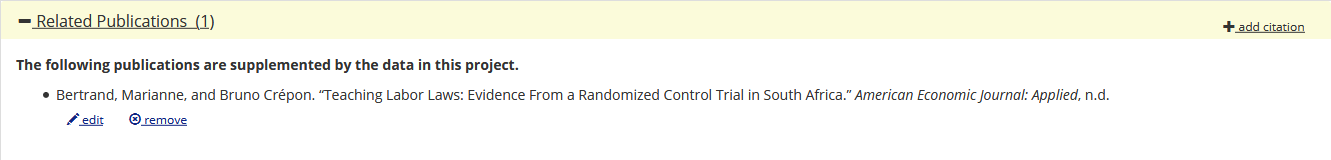
- Choose the “Import via DOI” button:
- Fill in the DOI (e.g.,
10.1257/rct.156-1.1) and press “Import”:
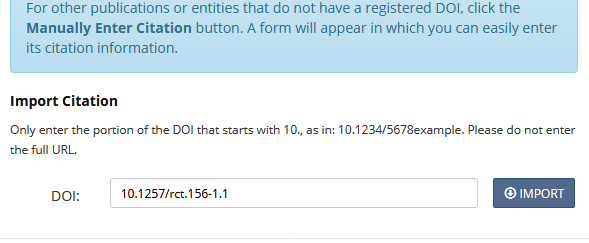
- Select “
is supplemented by” and press “Save and Apply”