Use of Docker for Reproducibility in Economics
Published:
In reproducibility verification, a common scenario is the author response “but-it-works-on-my-machine”. Finding common environments is important in such situations, to demonstrate that the error does arise, reproducibly, but also to share with the author the exact environment so that the issue can be fixed. Shipping around laptops probably isn’t the right solution. We illustrate how we addressed some of those cases using container technology throughout this post.
NOTE: There are probably still errors in this post. It will remain draft until the stream of comments has died down…
NOTE 2: Post updated with final publication dates for some articles.
What are containers
Containers are “implementations of operating system-level virtualization,” 1 typically on Linux. The most common version is provided by Docker, but several other implementations exist, such as Singularity. The use of containers as part of replication packages in economics is extremely low, and yet they have some advantages. This post will explore both pre-submission and post-publication uses of containers, as well as several shortcomings.
In a nutshell,
“A container is a standard unit of software that packages up code and all its dependencies so the application runs quickly and reliably from one computing environment to another.” 2
In particular, this means that all dependencies are handled, and there should be virtually no differences across time, users’ operating system, and software implementation in reproducing the outcomes of software.
Containers also isolate the computation from the host system, so that no user-specific settings (stuff you have configured over the past 10 years of working on a project) are carried through into the container, up to a point. That kind of isolation can also be achieved in various ways: for the cognescenti (also called geeks), chroot environments on Unix systems go back to the early 1980s 3, and “virtual machines” probably go back to 1960s IBM mainframes 4)
Containers and reproducibility
Containers can, but do not necessarily contribute to reproducibility and replicability. For an excellent introduction, see Boettinger (2015) 5. Once created, containers can (should) reliably reproduce the context and output they were designed to handle. Because containers typically use a plain-text script for their creation (so-called Dockerfile), they can contribute to transparency. However, containers can also be interactively created, or incorporate externally created packages, which may lead to some black-box actions that are not transparent. Thus, containers can generate reproducible output, but not be themselves reproducibly created. Furthermore, if containers rely on external resources – for instance, accessing the internet to download data from an API, or installed “latest” packages – the output generated may not be reproduced, though it may still be considered replicable.
Containers in computational social science
For the (social science) research environment, this should, in principle, handle many of the common problems that workarounds and READMEs list. In this post, we will explore several such cases, in which a computational issue was encountered, debugged leveraging both locally installed and cloud-based containers, and ultimately solved.
- Run R through a container
- Run Intel Fortran (or another complex setup) through container
- Run Gurobi through container with a dynamic license
- The Holy Grail in Economics: Run Stata through a container
The examples here are based on personal research experience as well as the experience when attempting to conduct reproducibility verifications on nearly 1,000 economics articles. They are by no means meant to be exhaustive or cover all the possible uses of containers. We should note that many of these examples are retro-actively ported to containers, and may not constitute the optimal setup. In our experience with 1,000 economics articles, as of November 2021, we have encountered only 1 (in words: one) that used any Docker in their submission to the AEA journals: Lamadon, Mogstad, and Setzler, “Imperfect Competition, Compensating Differentials and Rent Sharing in the U.S. Labor Market”,6 who leveraged 4 different CodeOcean compute capsules to support their analyses (we will return to CodeOcean later).
Running R, Julia, Python in Docker
Many of the examples one can find on the Internet sees container used in conjunction with open-source languages. For most statistical and other languages, in particular open-source languages, progress is continuous. In part what attracts researchers to these environments is the rich eco-system of add-on packages and libraries, most of which are available for free as well. However, combining versions of languages, packages, and dependencies to those packages can be a challenge. Use of these can sometimes be tedious - and not easily reproducible - because of a constantly evolving environment and very active development. Various methods exist to “pin” packages used (see renv, checkpoint and others in R, “environments”, pip freeze or conda env export in Python, Manifest.toml files and package environments in Julia). These rely, of course, not just on the ability to install the right packages, but also the right base software. In some cases, it can be challenging for users to install multiple base versions simultaneously on their computers - either because they work on multiple projects on their own at different times, or in collaborations with others. This is sometimes necessitated by changes that create breaks between versions (examples include changes in random number generators in Julia and certain breaks in R when version 4.0 was introduced). Programmatic solutions for this exist, of course, but add complexity to an often complex set of programs.
Thankfully, that same open-source community has also provided containers that lock in some of the base software versions, facilitating the simultaneous maintenance of multiple versions of the same software. We illustrate this with an example that we have used in some reproducibility cases with R. It addresses the particular use case of having multiple versions of R, but does not pin the particular package versions. See the tutorials for renv and checkpoint for even finer control.
A Really Simple Example with R
To start, we identify from instructions provided by the author both the version of R and the required packages. From the required packages, we use a standard setup program and add those libraries to the list of dependencies.7 We then leverage the R Docker images maintained by the Rocker group, and build a custom R Docker image that should have all dependencies as articulated by the authors:
Dockerfile:
FROM rocker/r-ver:4.0.1
COPY setup.R .
RUN Rscript setup.R
That’s already all there is to creating a Docker image! Of course, it now needs to be built:8
DOCKER_BUILDKIT=1 docker build . -t $MYHUBID/$MYIMG:$TAG
where I have previously configured some environment variables for ease of notation and reference:
TAG=v$(date +%F)
MYIMG=aer-9999-8888
MYHUBID=aeadataeditor
This locks in our R environment, at the time of building ($TAG). It is then straightforward to run the authors’ code, reliably, over and over again:
cd path/to/code
docker run -it --rm -v $(pwd)/subdir:/code -w /code $MYHUBID/${MYIMG}:$TAG Rscript main.R
Note that for each project, we can specify a different R version (rocker/r-ver:4.0.1), and we can even, for the same project, test the reproducibility with multiple R versions, by building multiple images (we might need to refine our tagging for that to work).
Furthermore, if we run into reproducibility discrepancies, we can share the resulting (public) Docker image and the “very simple” instructions with the authors. In doing so, we reproducibly share the failure to reproduce. We have done so, in fact, in several cases, though we will describe that particular type of interaction in the next section, in somewhat more challenging cases.
All we need is Docker as a software installation on our workstation. Or: in the cloud.
Running Docker the Cloud way
Numerous cloud computing providers facilitate running Docker. Some might call it an “app” that you are running, others may require you to boot up a Linux VM before running Docker, the mechanisms vary substantially. In many instances, you may need to worry about how to recover the results once the Docker image has finished running your code, and both data and code will likely need to be embedded in the Docker image, something not addressed here.
One innovation that multiple authors have recently pointed to is the July 2021 release of “Github Codespaces.” Subject to some conditions, anyone can easily run arbitrary Docker images in the cloud. We illustrate this using a reproducible environment to conduct a tutorial (readers might want to consult the tutorial for several other reasons as well).
The tutorial is programmed in R, leverages R-generated HTML slides (ioslides). Relevant for the point here is that the tutorial is intended to be conducted interactively. To do that, after having enabled Codespaces for the organization, we select and launch a codespace - essentially, a cloud-computer running our code.
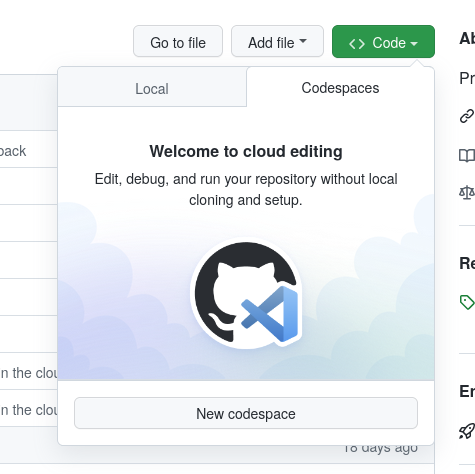
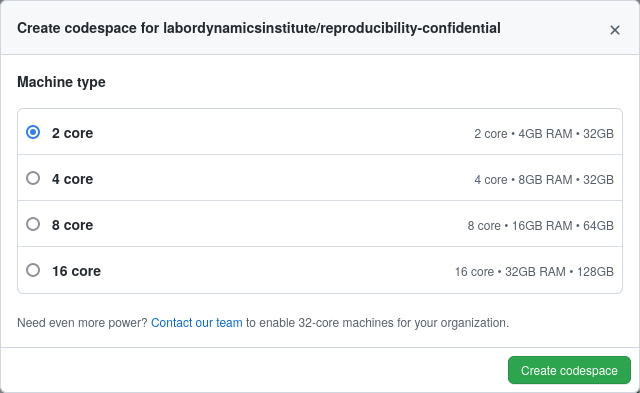
After choosing a size (which can go up to a 16-core/ 32GB RAM server), the system launches into a simple Linux shell, with the source git repository already checked out
Docker comes into play here because we want to use a Rstudio instance in the cloud, already set up the way we need it to be. The build script does most of the original legwork for us, and once in the cloud, all we need to do is to launch the pre-configured (reproducible!) Docker instance, loaded with our libraries and code:
docker run \
-e PASSWORD=testing \
-v $WORKSPACE:/home/rstudio \
--rm -p 8787:8787 $dockerspace/$repo
which we do straight from the cloud command line:
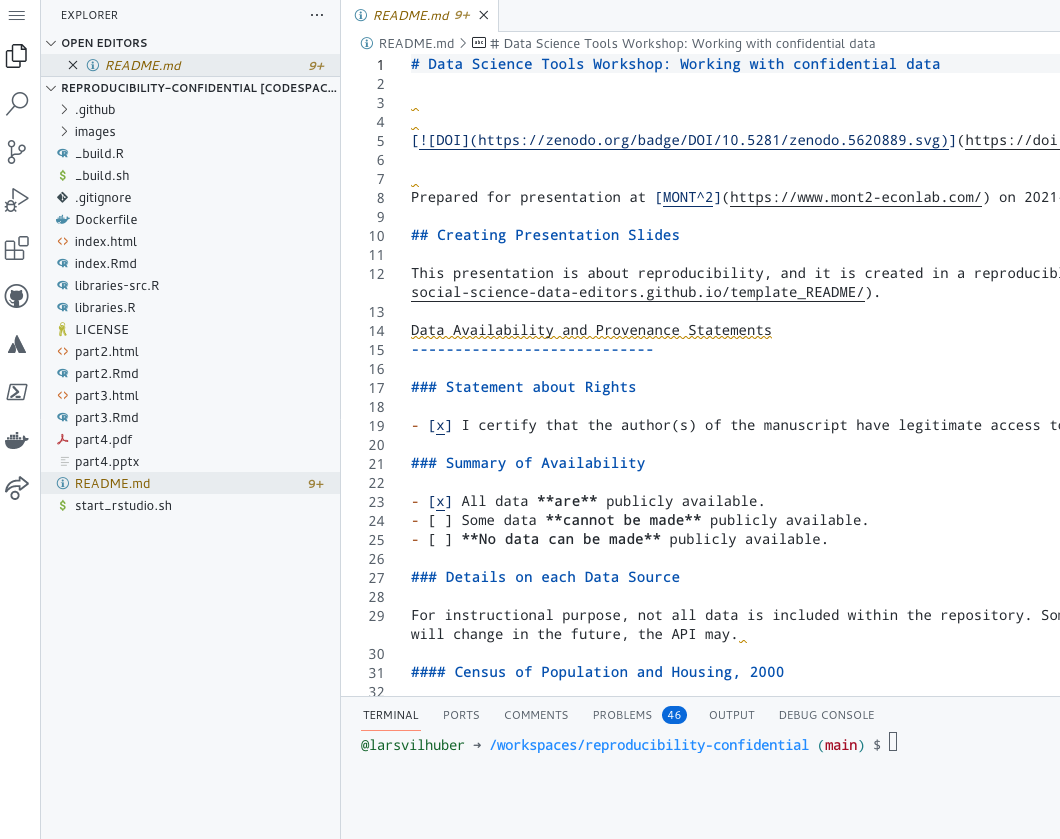
The docker command again downloads the image components from the Docker Hub, and runs the Rstudio instance:
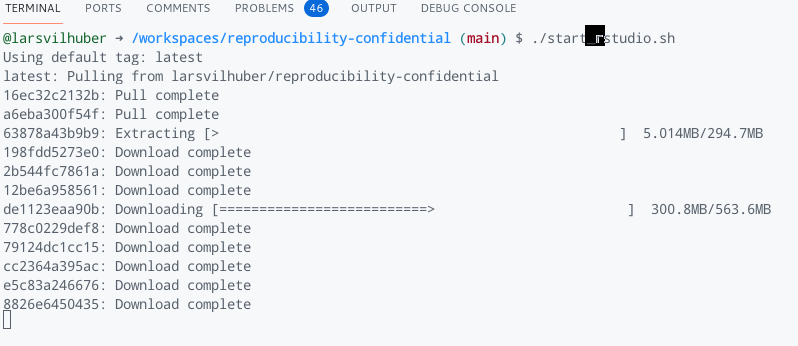
Codespaces detects that we are running a webserver within the Docker image, and prompts us to connect to it:
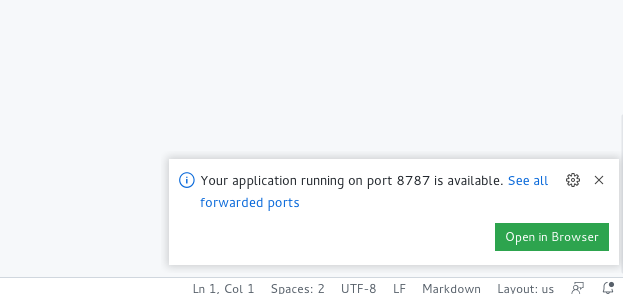
This is an example of reproducibility for interactive uses, but would apply in much the same way if all we wanted to do is run some R code repeatedly, or reproducibly. In fact, that is exactly what a separate functionality (Github Workflow/Actions) does to generate the slides for this particular tutorial, something left for next time (but see the workflow configuration file for the tutorial). We will come back to this later.
Compiling Fortran code through a container
Compilers are the workhorses for high-performance computing, and many economists will program compute-intensive routines in Fortran, then compile and run such code. Both open-source 9 and commercial10 are available. We describe two use cases for using Fortran in combination with Docker.
CE Fortran
In many cases, compilers require the presence of many separate libraries, and installation is traditionally complicated. The CE-Fortran project (“Computational Economics using Fortran”, Hans Fehr and Fabian Kindermann) trains and guides economists using open source GNU Fortran.
They have traditionally provided detailed installation instructions for all three operation systems, striving to make the development environment as comparable as possible across all three. More recently, the authors have started providing a Docker image to provide a consistent environment, see their Github page.
Nevertheless, some of the issues cannot be fully handled in a Docker image. For instance, while the environment in the base Docker image can create images, it cannot be used to provide an IDE or for manipulating images interactively. For this purpose, the authors provide a second, much larger Docker image that includes a workaround (a VNC server).
Intel Fortran
A second scenario is frequently encountered among the AEA’s replication packages. Many authors, particularly in macroeconomics, use Intel Fortran compilers. While installation instructions have been streamlined, they remain onerous: the most recent Windows installation instructions rely on a two installers, and have a dependency on Microsoft Visual Studio - a separate installer - which in turn has multiple additional dependencies. Furthermore, authors rarely use Makefiles, a way to compile code and address dependencies commonly used in the open source community, and only sometimes use Windows-specific Visual Studio Project files. The typical instruction, repeated dozens of times, looks like this:

with subsequent instructions to manually run the compiled code. Such instructions are inefficient, and potentially error prone.
A Fortran Docker Example
In reproducing a recent paper, we instead opted to streamline the process, making for a more efficient reproducibility check, and also – if the authors were to adopt this process – a more efficient development process.
- Using scripting tools, we extracted all of the
ifortcommands from the README PDF (34 lines) - We manually inserted the relevant directories, and added code to automatically run the compiled binary.
- The resulting script file (
run_all.sh) looks like this (inbashnotation - this could also be done as a Windowsbatfile):
# Code to run models
# Author: Lars Vilhuber
# Based on instructions in the paper's replication package
# NOTE: this should really be a Makefile!
# Capture base folder
BASE=$(pwd)
# Compiler options
IFORTOPTS=" -qopenmp"
# Figure 1A
cd $BASE/rep_agent_models/tc_model_nongrace
time ifort $IFORTOPTS rep_tc_delta_array.f90 rouwenhorst.f90 \
-o rep_tc_delta_array.exe && time ./rep_tc_delta_array.exe
cd $BASE/rep_agent_models/tc_model_grace
time ifort $IFORTOPTS rep_tc_grace_delta_array.f90 rouwenhorst.f90 \
-o rep_tc_grace_delta_array.exe && time ./rep_tc_grace_delta_array.exe
# and so on...
Simplicity
Once we had the script, we used the Intel OneAPI-hpckit (required 23GB of disk space, compared to the 24GB that the Windows install would have taken):
docker pull intel/oneapi-hpckit:2021.2-devel-ubuntu18.04
- Ran master script through the docker image
cd PATH/model_codes docker run --rm -it -w /code \ -v $(pwd):/code intel/oneapi-hpckit:2021.2-devel-ubuntu18.04 \ /code/run_all.sh > run_all.output.txt
This generated about 100 output files, without any further manual intervention. There is no separate installation process for the Intel compilers, a simple docker pull command allowed us to run all of the required files, within a few minutes.
Flexibility
When one of the compile steps required a larger memory size (25GB) than our workstation had available (23GB), we simply re-ran the following three steps on a general purpose server we had never before used for this purpose (but which already had Docker installed):
git pull (URL of the reproducibility repository)
cd PATH/model_codes
docker run --rm -it -w /code \
-v $(pwd):/code \
intel/oneapi-hpckit:2021.2-devel-ubuntu18.04 \
/code/run_all.sh > run_all.output.txt
(the docker pull command is automatically executed by the docker run command if the image is not locally available).
Thoughts
When using a container for the task of running these (fairly standard) types of structural models, we gained robustness and simplicity. Instead of a complex and manual Windows install specific to this type of project, we had a very simple way to run the author-provided code. The “transparency boundary” remains unchanged: the reliance on the proprietary (non-open source) Intel compiler, whether installed as a Windows installer, or obtained via a Intel-provided Docker image, is conceptually similar, and similarly opaque.
Running Docker the Easy Way
Of course, this may all seem overwhelming and complicated, as any new piece of software. Which is why several services are available that make this all a lot easier. These include CodeOcean, WholeTale, Binder, and others (see Konkol et al, 2020,11 for a broader overview). Behind the scenes, these use containers as well, sometimes built on the fly, but also able to archive the containers, ensuring persistence of what hopefully are reproducible containers.
We have started to use these services to enable reproducibility verification in an environment which allows us, when the time inevitably arrives, to share the exact environment with the authors in order to debug code. In contrast with the statement “Author, please use this Docker image to reproduce the issue”, the request “Author, please click on this URL to reproduce the issue” is much simpler. Several authors have been offered both options (no RCT…), and have generally preferred the option of an online easy interface to the ability to run Docker or Singularity locally.
And we have started to leverage the ability to also publish these reproducible artifacts. Recent examples include Rossi (2021)12 and DellaVigna and Pope (2021)13. In both cases, we were able to populate the CodeOcean capsule when we ran into reproducibility issues, sharing it (via a URL) with the authors, and solving the issue that both satisfied the AEA Data Editor, and is likely to satisfy future replicators as well.
We are working with the WholeTale on similar usage scenarios, and are open to working with authors’ choices whereever those may lead.
One key aspect here is worthwhile highlighting. In contrast to some other online computational platforms, for instance Google Colab, the ability to download (nearly) the entire reproducible environment, not just the code, and archive it, are key. That does not mean that they are automatically “fully reproducible”. They may still be missing some key elements that replicators will need to contribute, such as licenses.
Licenses
Most examples out there use open source software, not requiring licenses. This is an issue in economics, where the most common software used - Stata, Matlab - are proprietary and require licenses. Luckily, there are many ways to include licenses in Docker images at runtime, or, if using them internally within a research group, embedding licenses within the Docker image, as one would install a license in any “real” computer. The online systems mentioned above sometimes provide access to such licenses as well. We briefly illustrate first the easy, online way to incorporate licenses into container-based workflows, and then two “hard” ways.
An example using Stata, licenses, and the “Easy Way”
Bringing many of the issues alluded to earlier together, we have successfully leveraged this “toolkit” to improve the reproducibility and accessibility of Stata-based replication packages. Stata is used by the vast majority of economists, so being able to handle this scenario is important, but it also illustrates how licensed software can be used with containers.
The easiest way is to use an online system that provides such access. The first to our knowledge was CodeOcean. Simply choosing one of the configured Stata images will create a Docker image with Stata:
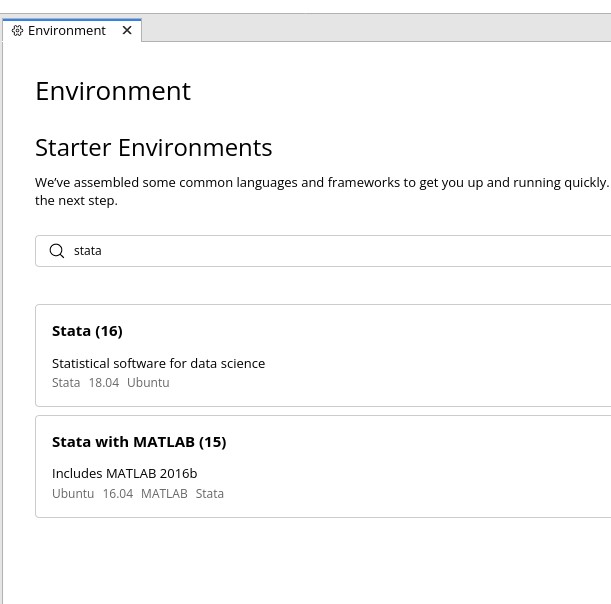
Hard-to-nail down failure to compute
This is the environment we used to work with the authors to create the reproducible package for DellaVigna and Pope (2021). We were able to work exclusively within CodeOcean’s environment, debugging a tenacious problem that wasn’t easy to diagnose. But we also wanted to make sure that the resulting replication package is not reliant on CodeOcean’s infrastructure. The authors debugged the code on CodeOcean (yielding the published capsule). We then verified the instructions to re-run the replication package downloaded from CodeOcean on a personal workstation, emulating a somewhat typical economist. We also populated the official replication package on openICPSR with the same code (https://doi.org/10.3886/E135221V1). This is strictly speaking not necessary, since CodeOcean is a trusted repository, and assigns DOIs, but for now, we make it easier to discover this way.
After downloading the CodeOcean capsule (57033059-76d7-422d-8301-d173e3520f07.zip), or equivalently, the openICPSR deposit, the following straightforward code (mostly documented in the CodeOcean-provided REPRODUCING.md) reproduces the paper’s outputs perfectly:
unzip 57033059-76d7-422d-8301-d173e3520f07.zip
cd environment/
# Need the Stata license
cp /path/to/statalicense/stata.lic.16 stata.lic
# Build the image - note, requires CodeOcean provided image. Could be replaced with the dataeditors/stata16 image
DOCKER_BUILDKIT=1 docker build . \
--tag 57033059-76d7-422d-8301-d173e3520f07
cd ..
# Ensure the script is runnable - this is not documented
chmod a+rx code/run
docker run --rm \
--workdir /code \
--volume "$PWD/environment/stata.lic":/usr/local/stata/stata.lic \
--volume "$PWD/data":/data \
--volume "$PWD/code":/code \
--volume "$PWD/results":/results \ 57033059-76d7-422d-8301-d173e3520f07 \
./run
Challenges
Big data
Since a container is meant to package up all necessary dependencies, it would seem logical that one could include all data. However, this breaks down when the data are large, even before getting into “big data” territory. The typical Docker image is no larger than 2-3 GB. Thus, the issues that plague reproducibility when data are large - delays in downloading or streaming data, availability of local space - are not solved by containers. That being said, satisfying the software dependencies is a first step in making sure that reproducibility can be achieved. Thus, a variant of the above way to run a Docker image could also be used in the presence of big data, by mounting the local storage directory for the data:
docker run --rm \
--workdir /code \
--volume "$PWD/data":/data \
--volume "/path/to/BIG/DATA:/bigdata \
--volume "$PWD/code":/code \
--volume "$PWD/results":/results \
researcherhub/papercontainer \
./run
Licenses not provided
Most of the “easy” systems are easy because they can accomodate a limited set of customizations. For instance, one can specify some post-install scripts on various platforms, but not choose an unsupported software image. But when the software requires additional licenses, a tension arises between publishing the successfully run image with an embedded license, and providing a template implementation that requires provision of a license.
Mathworks has gone the latter route, providing a Matlab Docker image without a license, and then providing instructions on how to provision the container with a valid license, for instance within a university’s internal network with a network server. Internally, WholeTale and CodeOcean use a similar mechanism behind the scenes.
Stata does not provide Docker image, but has allowed the AEA Data Editor to provide license-less Docker images for Stata. The instructions provided identify various ways a license can be provisioned to the image, so that code can be run:
docker run -it --rm \
-v ${STATALIC}:/usr/local/stata/stata.lic \
-v $(pwd)/code:/code \
-v $(pwd)/data:/data \
-v $(pwd)/results:/results \
$MYHUBID/${MYSTATAIMG}:${TAG}
In effect, users can simply use their own Stata license to enable a Stata Docker image. Again, WholeTale and CodeOcean use a similar mechanism behind the scenes, but the fact that any Stata user can re-execute such a Docker image with a valid Stata license is no more constraining that having installed Stata on their own computer.
In other circumstances, expiring licenses may be used. In a case where we needed to install Gurobi together with R, we used a new feature provided by Gurobi to provide “web licenses.” These can be disabled from a dashboard, and naturally expire after a while. Thus, our CodeOcean “postInstall” script installed Gurobi into a R container, then hard-coded a license key:
wget -v https://packages.gurobi.com/${GRB_SHORT_VERSION}/gurobi${GRB_VERSION}_linux64.tar.gz \
&& tar -xvf gurobi${GRB_VERSION}_linux64.tar.gz \
&& rm -f gurobi${GRB_VERSION}_linux64.tar.gz \
&& mv -f gurobi* gurobi \
&& rm -rf gurobi/linux64/docs
cd /opt/gurobi/linux64
#run the setup
python3.7 setup.py install
# Add the license key
# For replication purposes, this will need to be re-initialized, as post-publication, this key was disabled.
# You will need to provide your own.
echo "
WLSACCESSID=96388c95-2a7f-4195-88af-b167b077c019
WLSSECRET=ef818954-18ed-43f5-84b0-ace17641fd18
LICENSEID=6521234
" > /opt/gurobi/gurobi.lic
Once published, simply disabling the key on the Gurobi dashboard would prevent any unauthorized use of the license. Alternatively, of course, the method used for the Stata license above would also work outside of the CodeOcean environment, by using
docker run -it --rm \
-v gurobi.lic:/opt/gurobi/gurobi.lic \
-v $(pwd)/code:/code \
-v $(pwd)/data:/data \
-v $(pwd)/results:/results \
$MYHUBID/${MYGUROBIIMG}:${TAG}
Thus, generically, licensed software can very well be used when the proprietary software provider allows for redistribution of the binary portion of the software in the form of “opaque” containers; however, in many circumstances, the “easy” online providers of such container-based services can make it non-trivial to implement.
Complex interactions between software
Finally, as one example where limitations are hard to address ex-post, we highlight an example where researchers called a separately compiled Fortran binary from within Matlab. While each software could easily be run in its own container, it becomes non-trivial (but not impossible) to use multiple containers in this way.
The solution, as evidenced by the Gurobi example earlier, consists in installing one software into a container provided by the other. This, however, may lose the advantages of using pre-configured containers that avoid complex installation procedures, as in the case of Matlab and Intel compilers. An alternative (not tested) might consist in leveraging multi-stage builds, where entire portions are copied from one container into another. This is beyond the scope of the “easy” web-based solutions (though it may be on their development plan). However, precisely because a knowledgeable researcher can handle the (now much more complex) container creation, future users may not have to. By publishing the associated Dockerfiles, simple or more complex, future users do not need to reinvent the wheel - one of the purposes of the transparency that reproducibility fosters.
Continuous integration
Containers are also a tool for continuous integration - the software engineering best practice to test software at every step - but which can also be used to streamline research analyses. Consider the scenario where all data is available in a researcher’s Git repository (because of course, the researcher is using a Git repository). Containers can then be leveraged to create output - a research article, preliminary analyses, or a number of webpages. For instance, many researchers now use R in combination with Markdown to create “dynamic documents”. The (perenially incomplete) training manual for our undergraduate replicators is “built” this way, using the great bookdown package by Yihui Xie, using a simple preconfigured container:
- name: Run docker for R
uses: docker://larsvilhuber/ideahandbook:latest
with:
args: "/github/workspace/_build.sh publish"
(see the Github Action workflow file for full details)
Graphical utilities
Some of the “easy” online systems do not provide the native coding environment that many users are used to, for instance the Matlab or Stata desktop applications. This makes it harder, in fact, for such researchers to adapt to these tools. We regularly observe code or instructions that use visual inspection of the results, or leverage other features only available in a full graphical environment. This is less a concern for more recent (and typically open source) software, such as R (availability of Rstudio as a web service), Python, and Julia (Jupyter notebooks). However, WholeTale is trialling (with assistance from us) a combination of reproducible “tale” combined with a graphical user interface for Stata. The Matlab Docker image provides a graphical interface, as does one of the CE-Fortran docker images, so these are not fundamentally problems.
(Preliminary) Conclusion
Containers provide a way to share computational environments in an efficient way, to make such environments robust to changes in the underlying software, and to publish the fully transparent methods that lead to such containers. They are, however, underutilized in economics replication packages and by economics researchers. This document has provided a few scenarios that illustrate cases where the use of containers can help improve reproducibility.
Resources
So is this all rocket science? While I’m sure that the rocket scientists use containers, it isn’t really that complicated. Here is a (almost surely incomplete) list of a few resources to get you started:
Economics Articles and Replication Packages using Docker
- Rossi (JEL, 2021): “Forecasting in the presence of instabilities: How do we know whether models predict”12
- DellaVigna and Pope (American Economic Journal: Microeconomics, 2022) “Stability of Experimental Results: Forecasts and Evidence”13
- Bonhomme, Lamadon, and Manresa (Econometrica, 2019): “A distributional framework for matched employer-employee data”, Github repository
- Sergio Firpo, Vitor Possebom. 2018. “Synthetic Control Method: Inference, Sensitivity Analysis and Confidence Sets” Journal of Causal Inference 6.2, https://doi.org/10.1515/jci-2016-0026. Compute Capsule at CodeOcean
- Matias D. Cattaneo, Nicolás Idrobo, Rocío Titiunik. 2018. “A Practical Introduction to Regression Discontinuity Designs: Volume I” Compute Capsule at CodeOcean
Tutorials
- Grant McDermott’s EC 607 graduate course, Docker lesson
- Carpentries tutorial on Docker
- Installing Docker on a managed Windows system can be challenging, and my poor colleagues Michael Darisse and Leonel Borja Plaza have suffered through it for you to bring you this tutorial.
- Installing Docker on a Linux system, on the other hand, can be as simple as
yum install docker. - The command line can help on Mac as well:
brew install docker.
- Installing Docker on a Linux system, on the other hand, can be as simple as
Examples and Images
Tools
R-centric
Git-centric
Services
These are the services listed in Konkol et (2020)11 that use Docker
as well as more recent
and
- Gigantum (not tested)
as well as many cloud service providers.
Disclosures
We received a generous compute and storage quota from CodeOcean, a free license to use Stata 17 for one year in cloud applications from Stata, and a subaward on NSF grant 1541450 “CC*DNI DIBBS: Merging Science and Cyberinfrastructure Pathways: The Whole Tale” from the University of Illinois to evaluate the platform for the purpose of reproducibility verification. None of the sponsors have reviewed this preliminary assessment, or have had influence on any of the conclusions of this document. CodeOcean currently offers academic users a certain number of monthly free compute hours. WholeTale is free to use, though the Stata desktop functionality is not yet publicly available as of the writing of this document in November 2021.
Notes
-
Wikipedia. 2021. “List of Linux containers.” wikipedia.org/wiki/List_of_Linux_containers, accessed on 2021-07-07. ↩
-
Docker.com. 2021. “What is a Container.” docker.com/resources/what-container, accessed on 2021-07-07. ↩
-
Wikipedia. 2021a. “chroot”. https://en.wikipedia.org/w/index.php?title=Chroot&oldid=1044767727, accessed 2021-11-19. ↩
-
Wikipedia. 2021b. “Hardware virtualization”. https://en.wikipedia.org/w/index.php?title=Hardware_virtualization&oldid=1053629506, accessed 2021-11-19 ↩
-
Boettiger, Carl. “An Introduction to Docker for Reproducible Research.” ACM SIGOPS Operating Systems Review 49, no. 1 (January 20, 2015): 71–79. https://doi.org/10.1145/2723872.2723882. ↩
-
Lamadon, Thibaut, Magne Mogstad, and Bradley Setzler. 2022. “Imperfect Competition, Compensating Differentials and Rent Sharing in the U.S. Labor Market.” American Economic Review with Github repository and 1 2 3 4 compute capsules. ↩
-
Even without using the
checkpointpackage, we might use the MRAN snapshot server, but a faster method is provided by the binary packages provided by the Rstudio Public Package Manager used by the Rocker images. ↩ -
For additional steps, and complete instructions, see github.com/AEADataEditor/docker-r-starter ↩
-
GNU Fortran and various LLVM-based Fortran front-ends are available, see a list at fortran-lang.org/compilers/. ↩
-
Konkol, Markus, Daniel Nüst, and Laura Goulier. 2020. “Publishing Computational Research - a Review of Infrastructures for Reproducible and Transparent Scholarly Communication.” Research Integrity and Peer Review 5, no. 1 (July 14, 2020): 10. https://doi.org/10.1186/s41073-020-00095-y. ↩ ↩2
-
Rossi, Barbara. 2021. “Compute Capsule for: Forecasting in the presence of instabilities: How do we know whether models predict [Source Code].” CodeOcean. https://doi.org/10.24433/CO.7940775.v1, which accompanies her article in the Journal of Economic Literature (Rossi, 2021), and which is also archived in the more traditional location as Rossi, Barbara. 2021. “Data and Code for: Forecasting in the Presence of Instabilities.” American Economic Association [publisher], Inter-university Consortium for Political and Social Research [distributor]. https://doi.org/10.3886/E147225V1 ↩ ↩2
-
DellaVigna, Stefano and Devin Pope. 2021. “Compute Capsule for: Stability of Experimental Results: Forecasts and Evidence” [Source Code]. CodeOcean. https://doi.org/10.24433/CO.0687784.v1, which accompanies a 2022 article in American Economic Journal: Microeconomics, and which is also archived in the more traditional location as DellaVigna, Stefano, and Devin Pope. 2021. “Data and Code for: Stability of Experimental Results: Forecasts and Evidence.” American Economic Association [publisher], Inter-university Consortium for Political and Social Research [distributor]. https://doi.org/10.3886/E135221V1 ↩ ↩2

