Preparing your files for verification
Published:
Published:
This document describes how to best prepare a replication package for an AEA journal. Much of the guidance here is not specific to our journals - the document links to other websites for tutorials, best practices, etc. The best moment to do the preparation described here is, in fact, when you start the project, not once you have had your manuscript conditionally accepted. However, all steps here can and have been successfully performed at the point of conditional acceptance.
The Data Editor of the American Economic Association endorses DCAS, the Data and Code Availability Standard v1.0. The AEA’s data and code availability policy is compatible with DCAS.
Key principle: Computational empathy
- Keep in mind: The replication package is meant to be run by others, who have none of the setup, packages, and data that the original author might have, on computers that may not run the same operating system.
- Treat the replication package as one of the methods to convey the methods that lead to your manuscript’s conclusions. Consider it a teaching tool, targeting young graduate students who may not be in your field.
Describing the contents of your replication package
Every replication package requires a document outlining where the data comes from, what data is provided, what requirements are needed to run the code in the replication package, how to run the code, what results to expect, and where to find the results. This is conventionally called the “README”.
- The AEA requires that the README contain a number of information elements. A convenient way to ensure that these elements are present is to use the template README for social science replication packages v1.1; however, you are free to provide this information in a format of your choice as well.

The following information is required in the README (unless a modifier indicates otherwise):
- Data Availability and Provenance Statements
- Statement about Rights
- License for Data (optional, but recommended)
- Details on each Data Source
- Dataset list
- Computational requirements
- Software Requirements
- Controlled Randomness (as necessary)
- Memory, Runtime, and Storage Requirements
- Description of programs/code
- License for Code (Optional, but recommended)
- Instructions to Replicators
- Details (as necessary)
- List of tables and programs
- References (Optional, but recommended)
Some more information is provided below, a full discussion is available at the template README for social science replication packages.
The README should be in a format that is easily readable online, such as a PDF or a TXT file. You can provide a Word or LaTeX source for the README as well, but that is not required.
Provide the README as part of your replication package, ideally in the root directory.
Data Citations
All manuscripts will be checked for data citations. If you have not done so, now is the time to add them to your manuscript.
What to cite
All datasets actively used in your replication package that are not your own must be cited. Just as with any other literature, this serves to properly identify the provenance of the information you use, and attribution of credit to the original creator. Data you created should also be cited, either by pointing to a data deposit you made elsewhere, or by pointing to your own (forthcoming) data deposit at the AEA. Data you collected should be cited with the original location where you obtained the data, even if you provide a copy in your data deposit.
- AEA Sample References
- Guidance for data citations
- Guidance on how to cite the data you collected
- Some additional considerations
Where to cite
Every citation has an in-text entry (Smith (2020) or (Smith, 2020)), and a list entry in the Reference section (see the Chicago Manual of Style Quick Guide). This is true for data citations as well.
- Your data citations should be listed in the Reference section of the manuscript.
In rare circumstances, when there are too many data citations to accomodate succinctly, data citations for sources can be deferred to an online data appendix. In all cases, for clarity, all data citations should also appear in the README, including with a separate Reference section.
Data citations and Data Availability Statements
In many cases, the data citation for a public-use dataset has a unique URL that is sufficient for downloading the dataset:
Bureau of Labor Statistics. 2000–2010. “Current Employment Statistics: Colorado, Total Nonfarm, Seasonally adjusted - SMS08000000000000001.” United States Department of Labor. http://data.bls.gov/cgi-bin/surveymost?sm+08 (accessed February 9, 2011).
However, in many other cases where the URL does not lead directly to the dataset, a separate “Data Availability Statement” needs to be provided, as part of the README. A simple data citation may not sufficient. See general guidance on data availability statements, and some examples in the template README.
Code
It should be obvious that your replication package should contain all code used to generate the results in your manuscript.
Code to include
This includes
- code to read in raw data,
- code to clean and process data,
- code to analyze data and estimate models, and
- code to generate tables and figures.
This applies even to
- maps and other complex visualizations (guidance)
- web scraping code
- code that processes confidential data (see below)
- code that anonymizes data
- code that prepares surveys and runs experiments (guidance)
In all cases, it is possible that “code” is not a program, but human-readable instructions on how to perform a manipulation.
- For instance, despite the availability of scriptable GIS software, many economists prefer to use GUI-based GIS software, and manually construct maps.
- For instance, many websites providing data have only an interactive process to acquire or request data, and no single download link. In such cases, detailed instructions on how to acquire the data may be necessary (see https://doi.org/10.5281/zenodo.10983009 for an example). We do encourage authors to use APIs where available.
How to include
Code should be included in its “native” form, i.e., the format that the software expects to read it in. For instance, Stata code should be in .do files, R code in .R files, Qualtrics surveys in .qsf files, etc.
No manual modifications
The replication package should reproduce the tables and figures, as well as any in-text numbers, by running code without manual intervention.
The only exception to this rule is a single change to set a small number of program and data directory paths.
While running a small number of distinct programs separately is acceptable (in some cases even desirable), it is not acceptable to require replicators to manually enter numbers, or configure parameter files, in order to reproduce the tables and figures in the paper. Nevertheless, it should be clear from the manuscript and the code how a replicator might deviate from the tables and figures in the paper.
Data structure of a replication package
The AEA uses the openICPSR platform for replication packages. The platform allows users to download complete “deposits”, or only subdirectories thereof. However, deposits of replication packages at other trusted repositories are also accepted, as long as they satisfy the requirements described here.
Users must not upload ZIP packages as files - rather, ZIP files can be used to structure code and data, but should be unzipped on the platform (“import from ZIP”). The exception is when there are more than 1,000 files in the repository.
The code and data should run as downloaded from the repository, without further manual modifications (creating empty subdirectories programmatically is acceptable). Because code tends to be small, but data can be large, we strongly advise to not commingle data and code - interested researchers can download the code directory by itself if they wish, without also downloading a potentially very large data directory.
A simple template might be
README.pdf
data/
raw/
cps0001.dat
analysis/
combined_data.dta
combined_data.csv
combined_data_codebook.pdf
code/
01_create/
01_readcps.R
02_readfred.R
02_analysis/
01_table1-5.R
02_figures1-4.R
results/
table1.tex
table2.tex
...
figure1.pdf
figure2.pdf
Structure in the presence of more than 1,000 files
ICPSR cannot accept deposits with more than 1,000 files. Therefore, we relax the rule that all data and code must be unzipped, though we still insist on the “smallest possible configuration”.
In most cases, it is a particular directory that is the primary culprit. Say you have
Structure-pre:
/code: 20 files
/data/
src1/: 25 files
src2/: 101 files
src3/: 3,000 files
then the ideal structure, taking into account the 1,000 file limit, would be:
Structure-post:
/code: 20 files
/data/
src1/: 25 files
src2/: 101 files
src3.zip: = 1 file, containing 3,000 files
(src3/ and its 3,000 files have been removed!)
Your README should provide instructions to the replicators how to recover the fully unzipped structure (there are cross-platform differences in unzipping, so be precise about the final structure, rather than the method of getting there).
Alternatively, the code can handle the unzipping - optional, but more robust.
Once you’ve adjusted that, zip up the whole structure (so a ZIP file that has inside it another zip file, plus the /code, /data/src1/, and /data/src2/ directories), and “Import from ZIP” when uploading to ICPSR.
- See also a similar entry to our FAQ.
Structure in the presence of confidential (unpublished) data
When the replication package relies on confidential data that cannot be shared, or is shared under different conditions, authors will have to
- prepare a confidential (partial) replication package
- this would contain the contents of
data/confidentialand possiblydata/conf_analysisfrom the example below.
- this would contain the contents of
- preserve (archive) the confidential replication package
- If the data cannot be removed from a secure enclave, they should nevertheless be archived wherever the confidential data are kept (see this FAQ)
- If the data can be shared, but are subject to access restrictions, follow this guide on creating a separate data deposit and, when creating the restricted deposit at ICPSR, follow these instructions on how to do so.
- prepare a non-confidential replication package that contains all code, and any data that is not subject to publication controls
- this would contain the contents of
data/raw,data/analysis,code/, and for reference,results/from the example below.
- this would contain the contents of
- ensure that replicators have detailed instructions on how to combine the two packages
- specify which (if any) of the results in their paper can be reproduced without the confidential data.
Clearly separate the restricted from the open-access data, both in terms of the raw data as well as the processed data:
data/
raw/
cps0001.dat
confidential/
ssa.csv
conf_analysis/
confidential_combined.dta
Keep in mind that you may be able to provide a subset of your replication package privately to the AEA Data Editor, see the Sharing restricted-access data with the AEA Data Editor page, or that you might be able to create a separate data deposit with a more limited license.
Authors might want to investigate the possibility of providing “fake” or “simulated” data that might allow replicators to run code, without necessarily obtaining meaningful results (functionality test).
- Sharing restricted-access data with the AEA Data Editor
- The Social Science Editors’ FAQ describes a related issue.
Creating a separate data deposit
In situations where the data has a different license, or where the data you include has value separately from your replication package, you can create a separate data deposit, and link it/reference it/cite it.
- Creating a separate data deposit
- Guidelines for depositing at other repositories
- Special case: PSID
- Special case: confidential data
Considering the replicator
The replicator of your package is likely to be less qualified than you are. After all, you are publishing something novel.
You should assume
- that the replicator has basic knowledge in how to run your software package, if the software is commonly used in economics
- Stata, Matlab, some others are commonly used
- Compiled or new computer languages are much less likely to be widely used, even if they are used in your subdiscipline
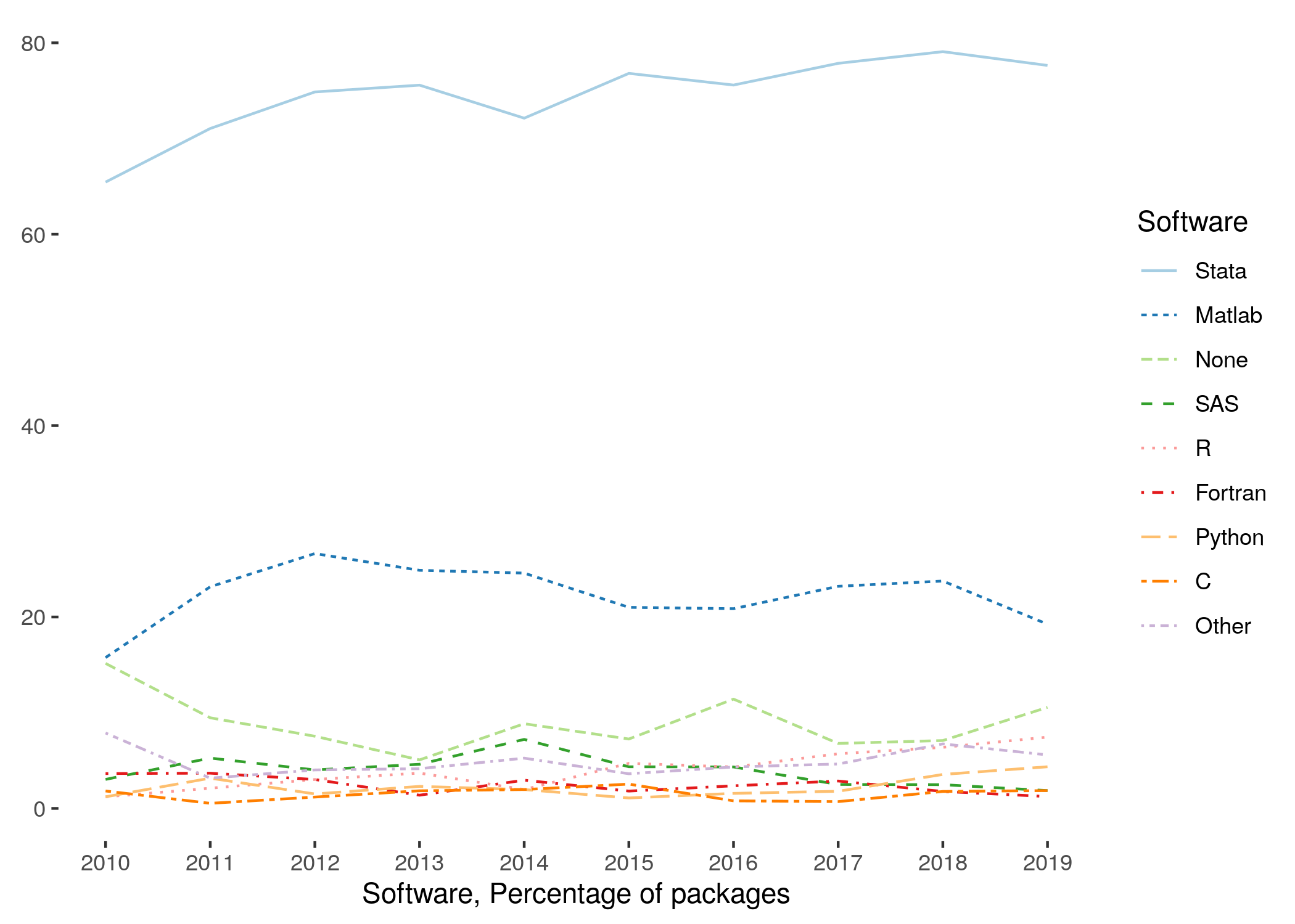
You can assume
- that the replicator can manipulate a top-level configuration file
- for instance, to set a base directory
- but not setting a base directory at the top of 25 different files
You should NOT assume
- that the replicator will use the same type of operating system
- describe any hard requirements, but do not impose any fake requirements
- most Stata, Matlab, SAS, R, Python, etc. can run on any operating system, unless you hard-code platform-specific commands into your code
- that the replicator has any of your packages/modules/etc. installed
- provide a setup program to install these (not manual instructions).
- provide copies of such packages/modules when the package repository does not allow you to specify a version
- provide a container/ Docker image/ VM that comprises all the necessary software and libraries
- that the replicator will run the software the same way you do
- some software can be run in different ways (interactive, batch, etc.) and may behave differently depending on how it s run.
- For instance, Stata will change the working directory to that of the program being run in batch mode, but not if running interactively
- For instance, running R code using Rstudio may behave differently than running it with
rscript
For less frequently used software, provide a URL where the software can be obtained.
- essentially, if not listed in the figure above, provide information on how to obtain software
- if using commercial compilers, we also suggest to compile your code using open-source or free compilers (including any free performance packages, such as Intel MKL), even if the resulting code is not the most efficient.
- as of 2021, the AEA Data Editor has access to the software on this list, and any open-source (free) software that can be installed on Windows, Linux, and macOS.
Choosing a license
By default, the openICPSR deposit attributes a Creative Commons Attribution 4.0 International Public License to your deposit, but you can choose a different license. If you do, you must add a file called “LICENSE.txt” (by convention capitalized) to the deposit.
- See our Licensing guidance for more details.
Re-run your replication package
Ideally, once you have prepared your replication package, you should re-run the code again, in a clean environment, possibly a fresh computer, to ensure that (a) the package is, in fact, reproducible with minimal interaction (b) the results are numerically identical.
- Wherever possible, we strongly encourage running in batch (non-interactive) mode.
Preparing to upload
Once you are done preparing your replication package, you should upload it:
- if you have received a conditional acceptance, your replication package must be in a trusted repository. The default trusted repository is the AEA Data and Code Repository. Other trusted repositories may be acceptable (see list), but replication packages should meet the display guidelines.
- if you have confidential data that you want to transmit to the AEA Data Editor but do not want published, communicate with the AEA Data Editor directly (see this FAQ and Sharing restricted-access data with the AEA Data Editor).
- if you have received instructions during the revise-and-resubmit process to have a reproducibility check conducted, you may use the AEA Data and Code Repository, but other methods are also acceptable. Do not forget, however, that once the paper is accepted, it must be made available on a trusted repository - other methods are then no longer acceptable.
Final checklist
Before proceeding, do check:
- you have prepared a README that provides all the relevant information, as per the README template
- your manuscript includes data citations
- your data and code deposit contains all code, including code to read in raw data, even when the data cannot be provided.
- your replication package has been re-executed, and reproduces the tables and figures in your manuscript faithfully.
Assistance
If, in the process of preparing your replication package, you have questions that are not addressed by these guidelines, please contact us:
- We are on BlueSky, Mastodon.
- Contact us via the AEA’s Contact form
- Reach out to the editor handling your paper, who knows how to reach us (but might also have the answer).
- Reach out to us directly (you know how to find us).
Next step
If you are ready, you can proceed to upload to the AEA Data and Code Repository (or your chosen alternate trusted repository).
